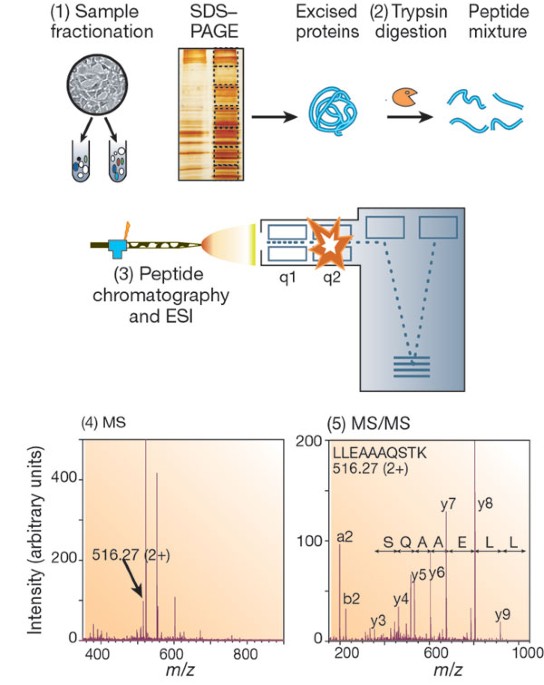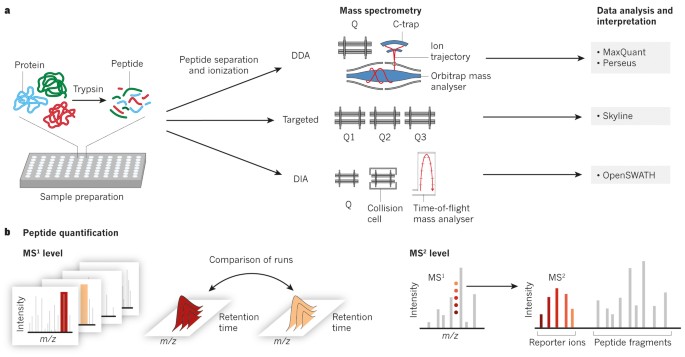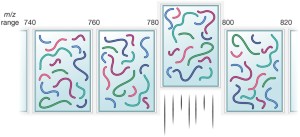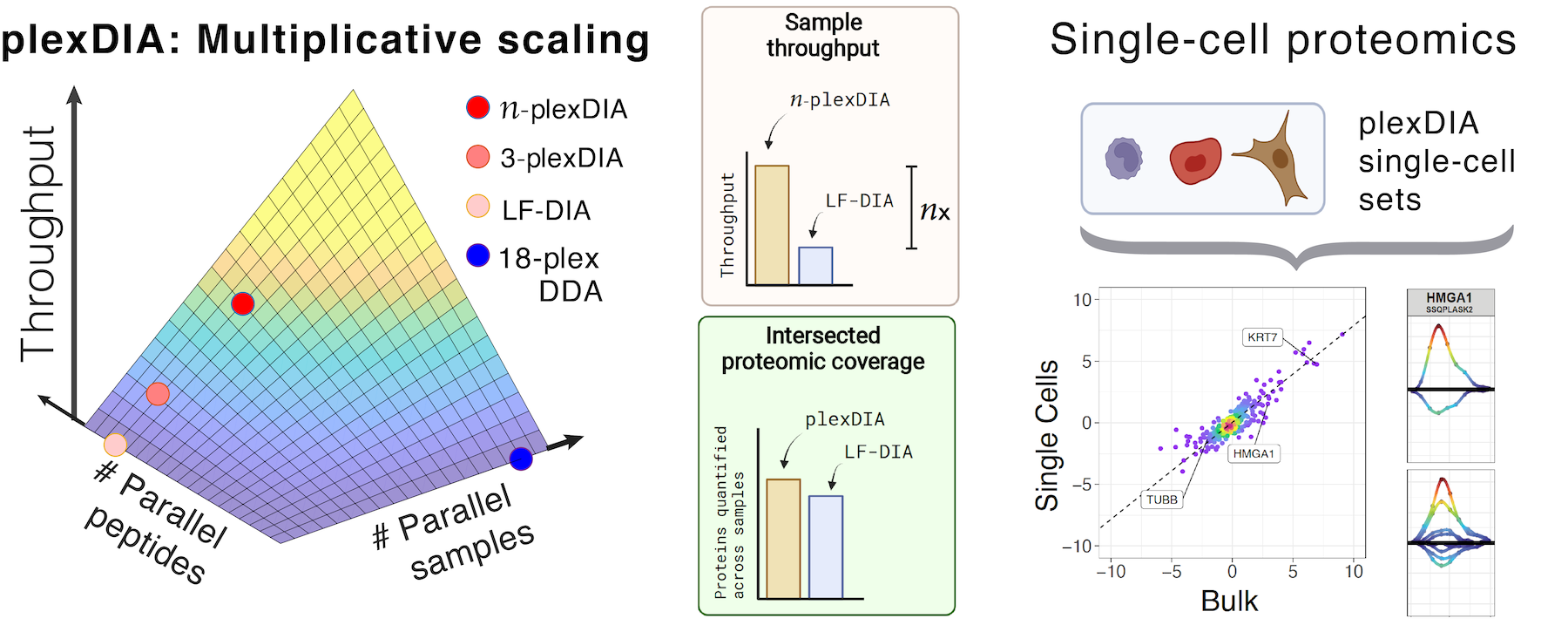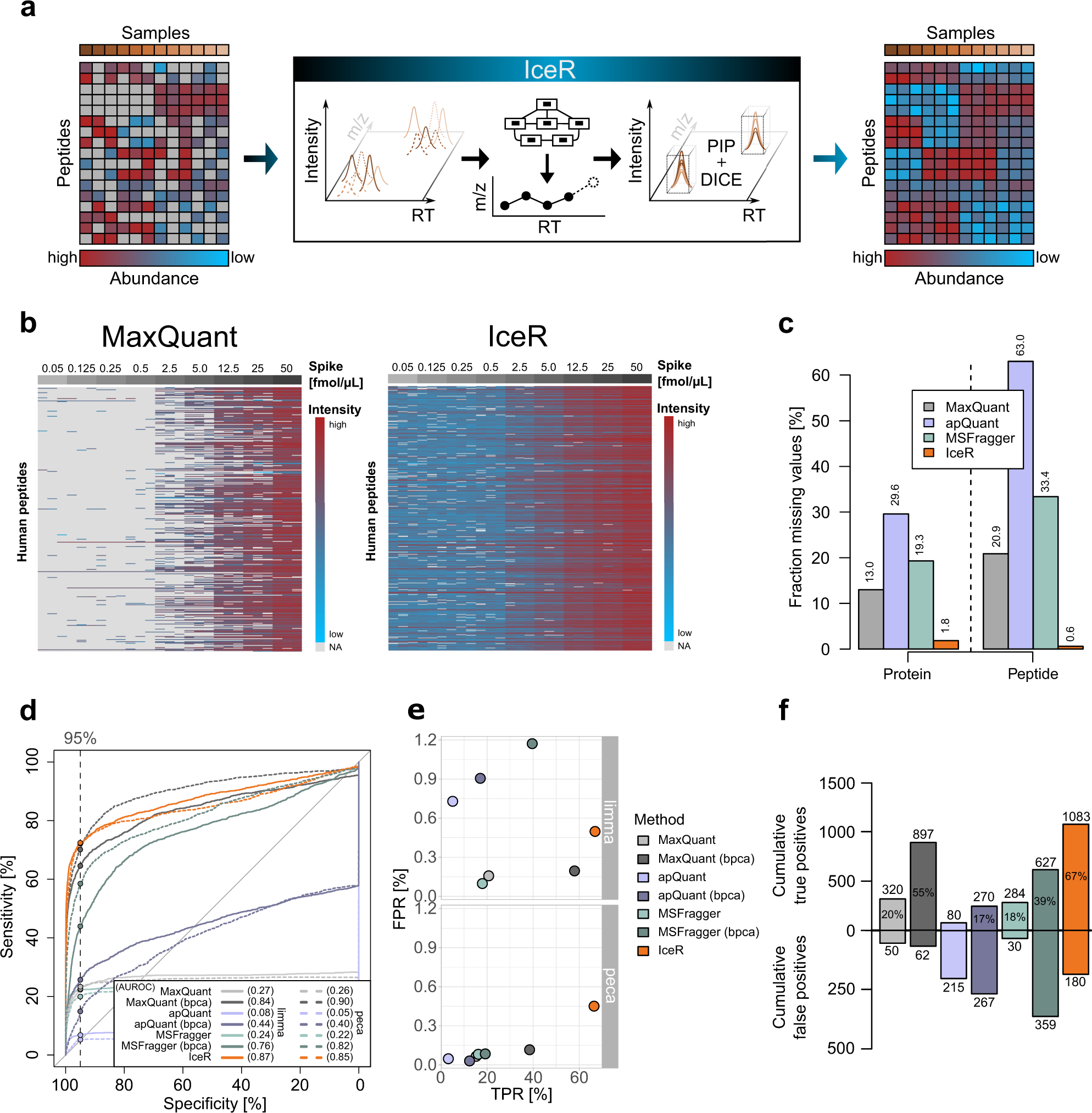
IceR improves proteome coverage and data completeness in global and single-cell proteomics | Nature Communications
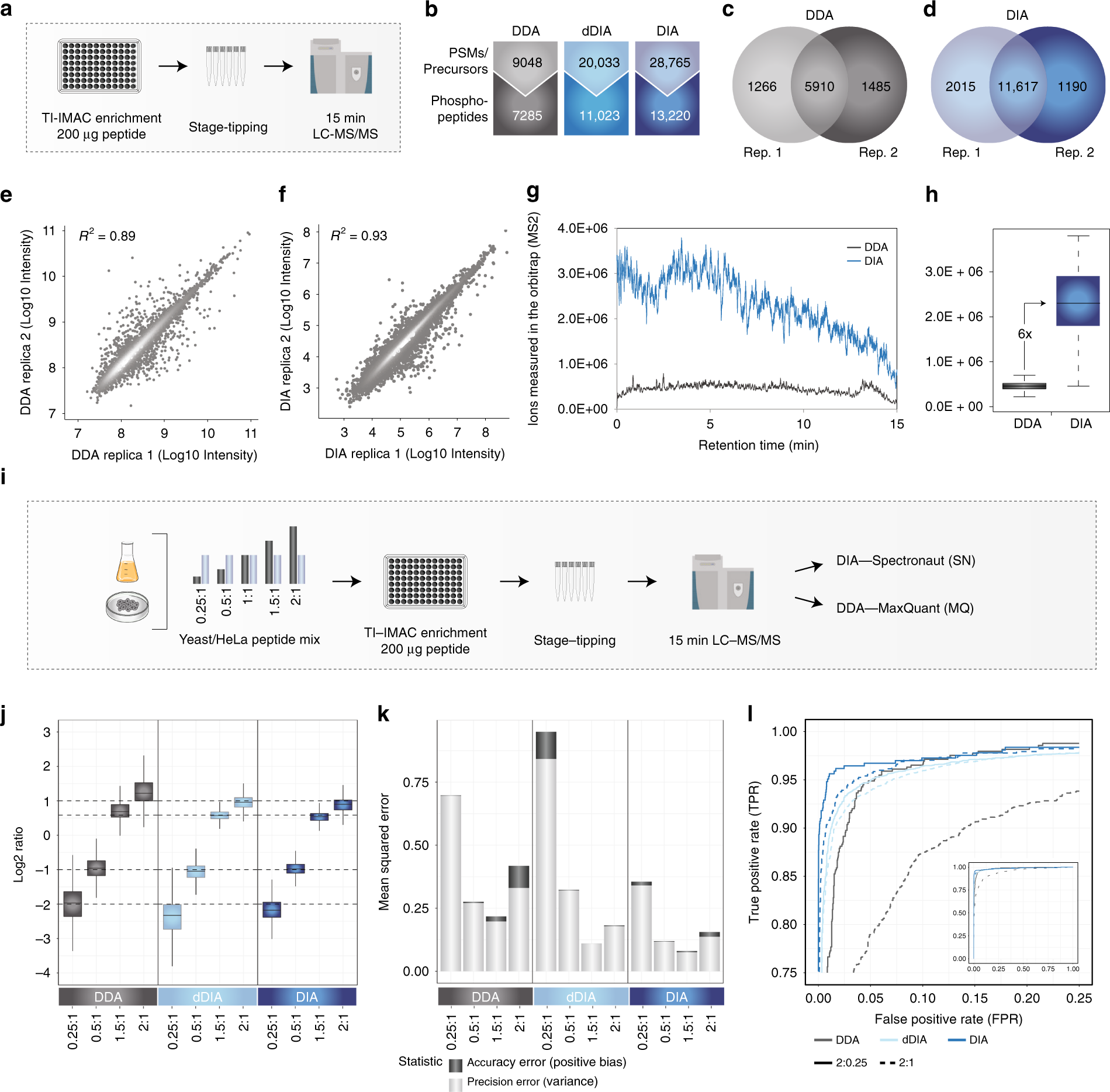
Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries | Nature Communications
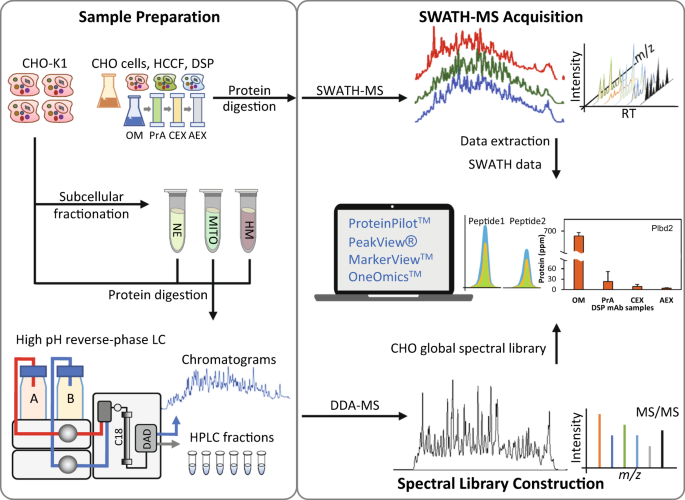
A comprehensive CHO SWATH-MS spectral library for robust quantitative profiling of 10,000 proteins | Scientific Data
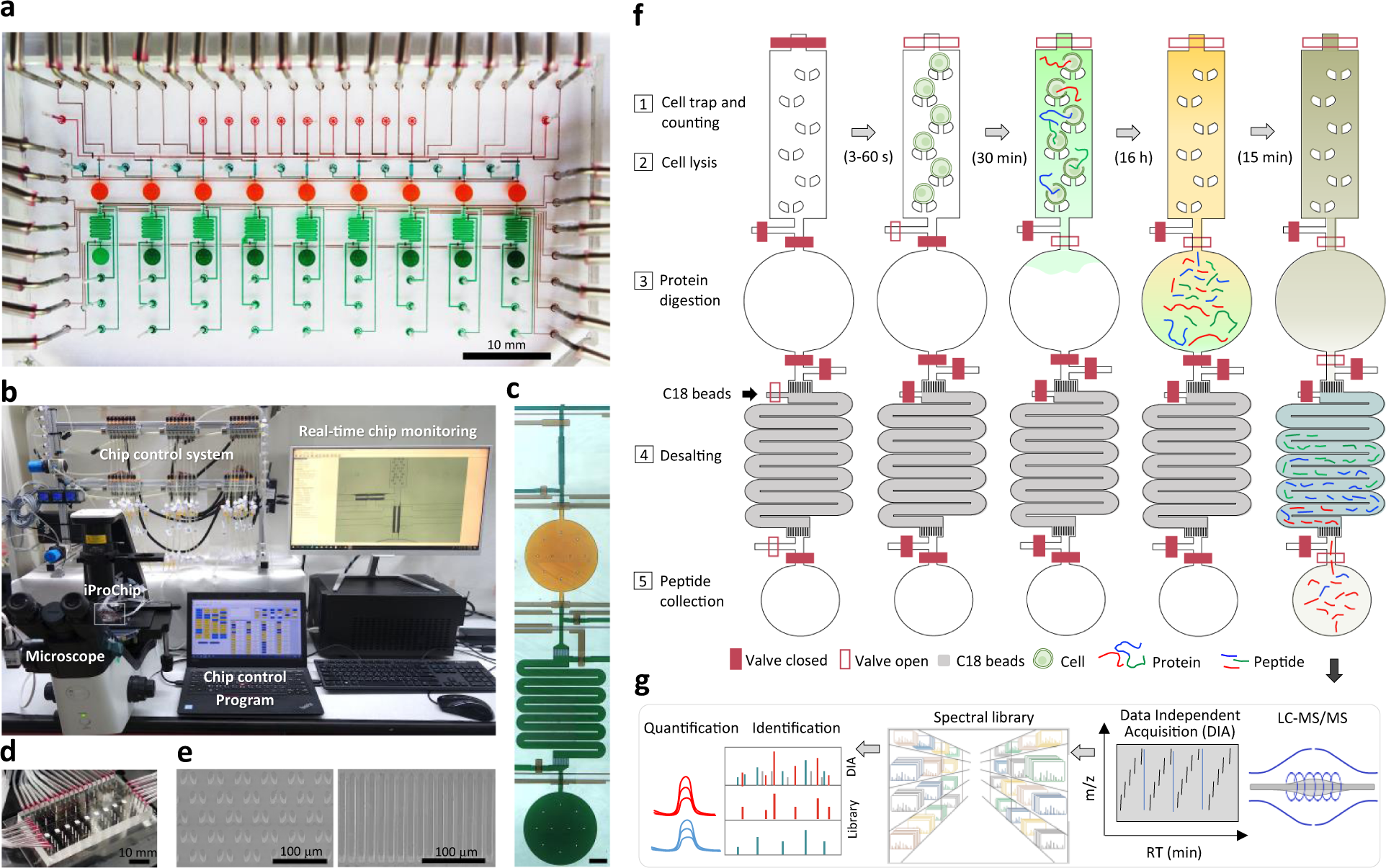
Streamlined single-cell proteomics by an integrated microfluidic chip and data-independent acquisition mass spectrometry | Nature Communications
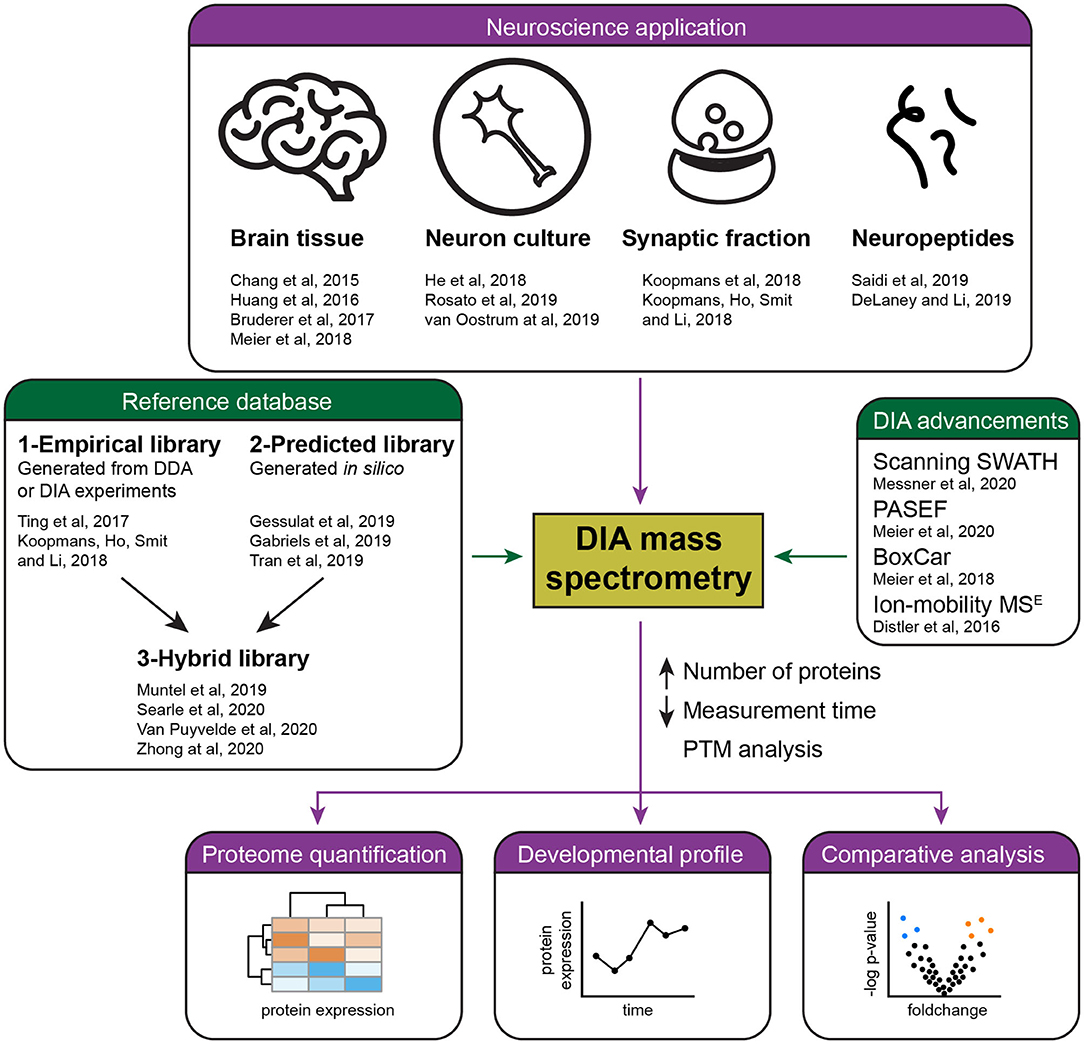
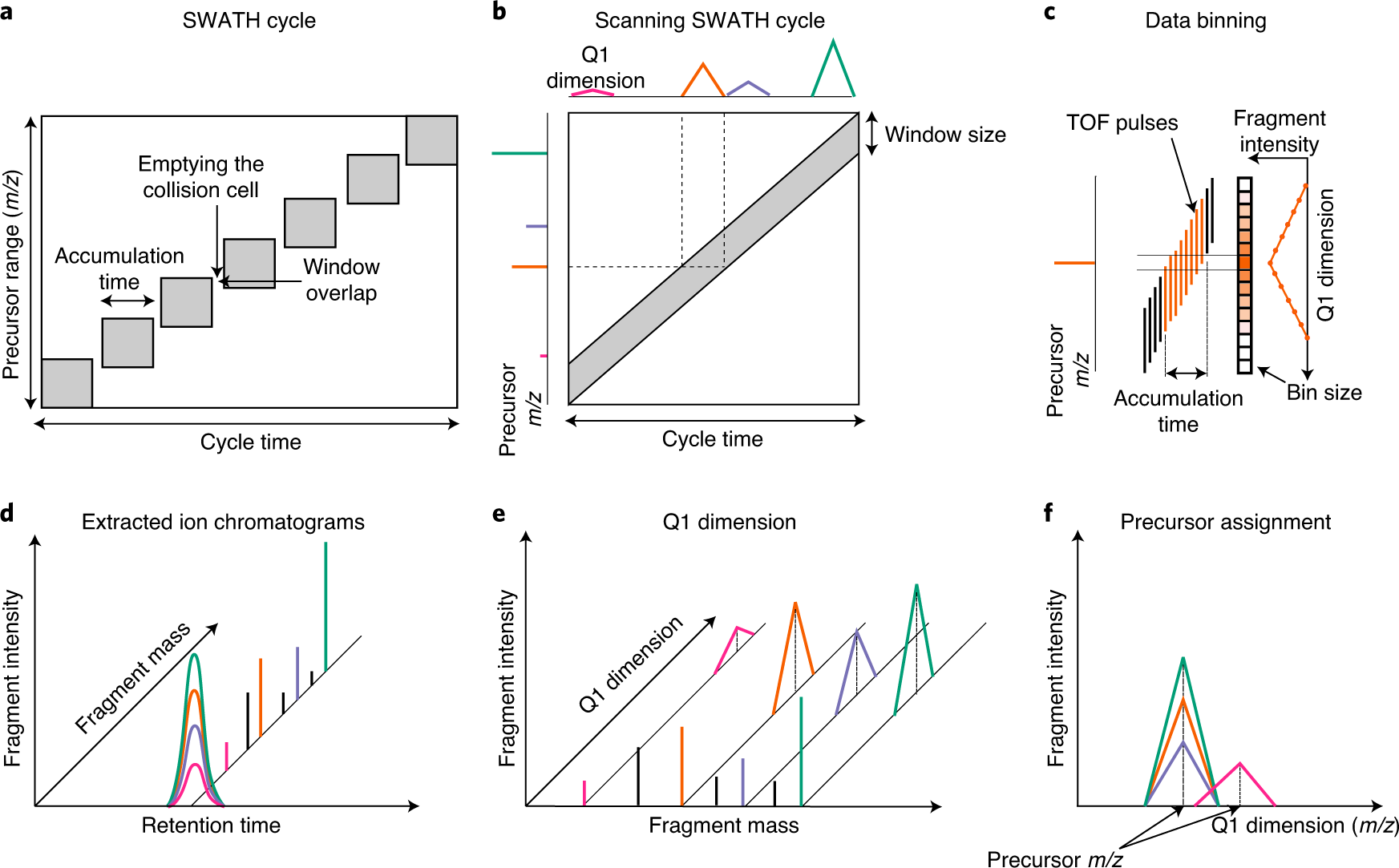
Frontiers | Recent Developments in Data Independent Acquisition (DIA) Mass Spectrometry: Application of Quantitative Analysis of the Brain Proteome

Group-DIA: analyzing multiple data-independent acquisition mass spectrometry data files | Nature Methods
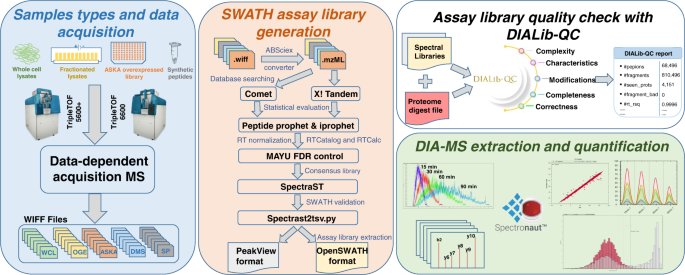
A comprehensive spectral assay library to quantify the Escherichia coli proteome by DIA/SWATH-MS | Scientific Data
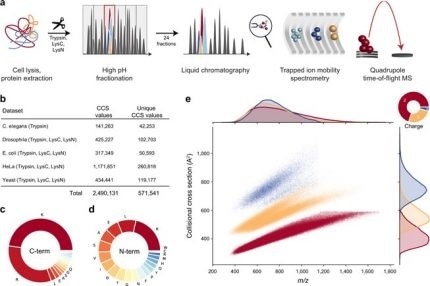
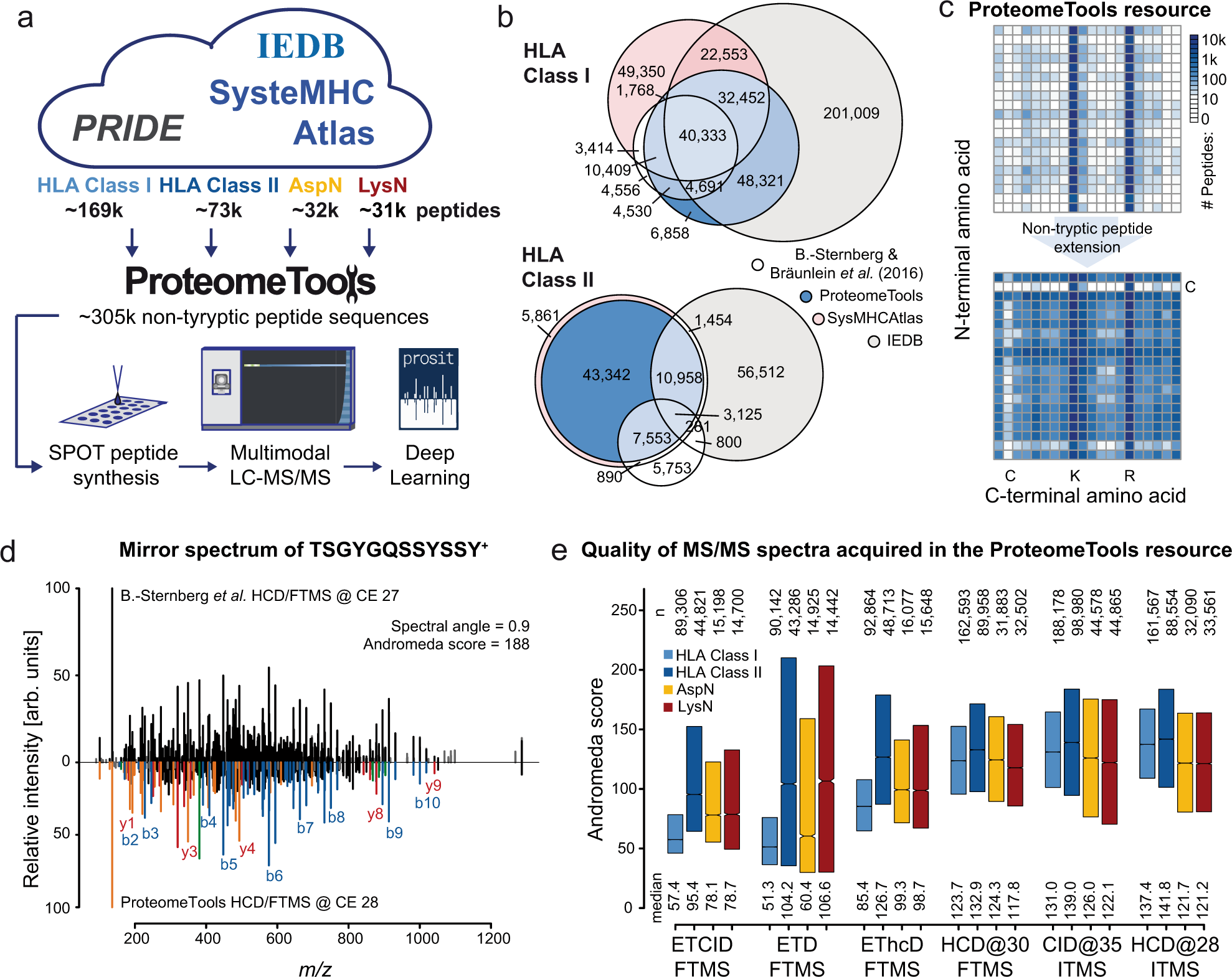
New Nature Communications publication by Mann & Theis Groups harnesses the benefits of large-scale peptide collisional cross section (CCS) measurements and deep learning for 4D-proteomics

Technical comparison of DDA and different types of DIA in a biological... | Download Scientific Diagram
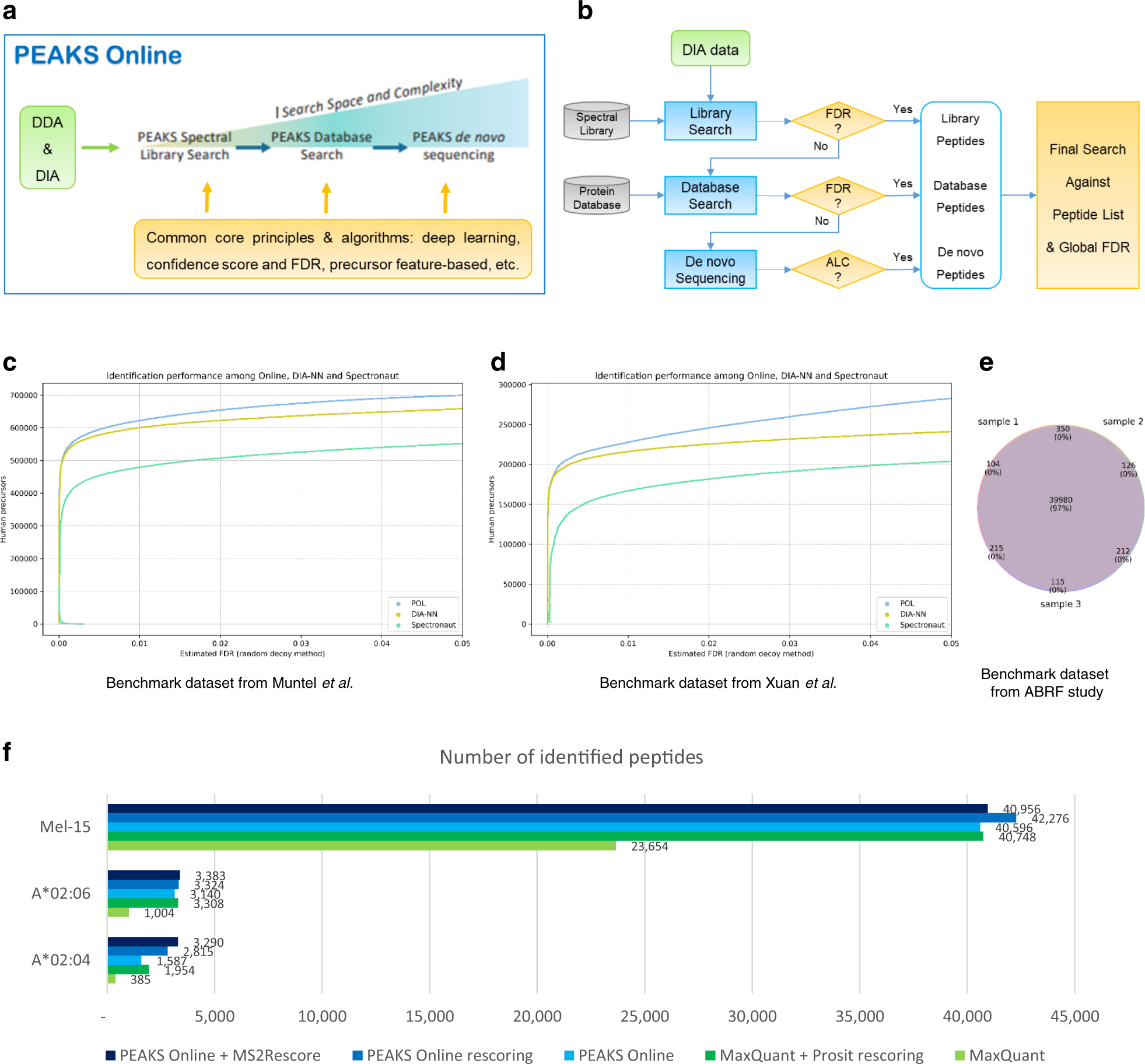
A streamlined platform for analyzing tera-scale DDA and DIA mass spectrometry data enables highly sensitive immunopeptidomics | Nature Communications
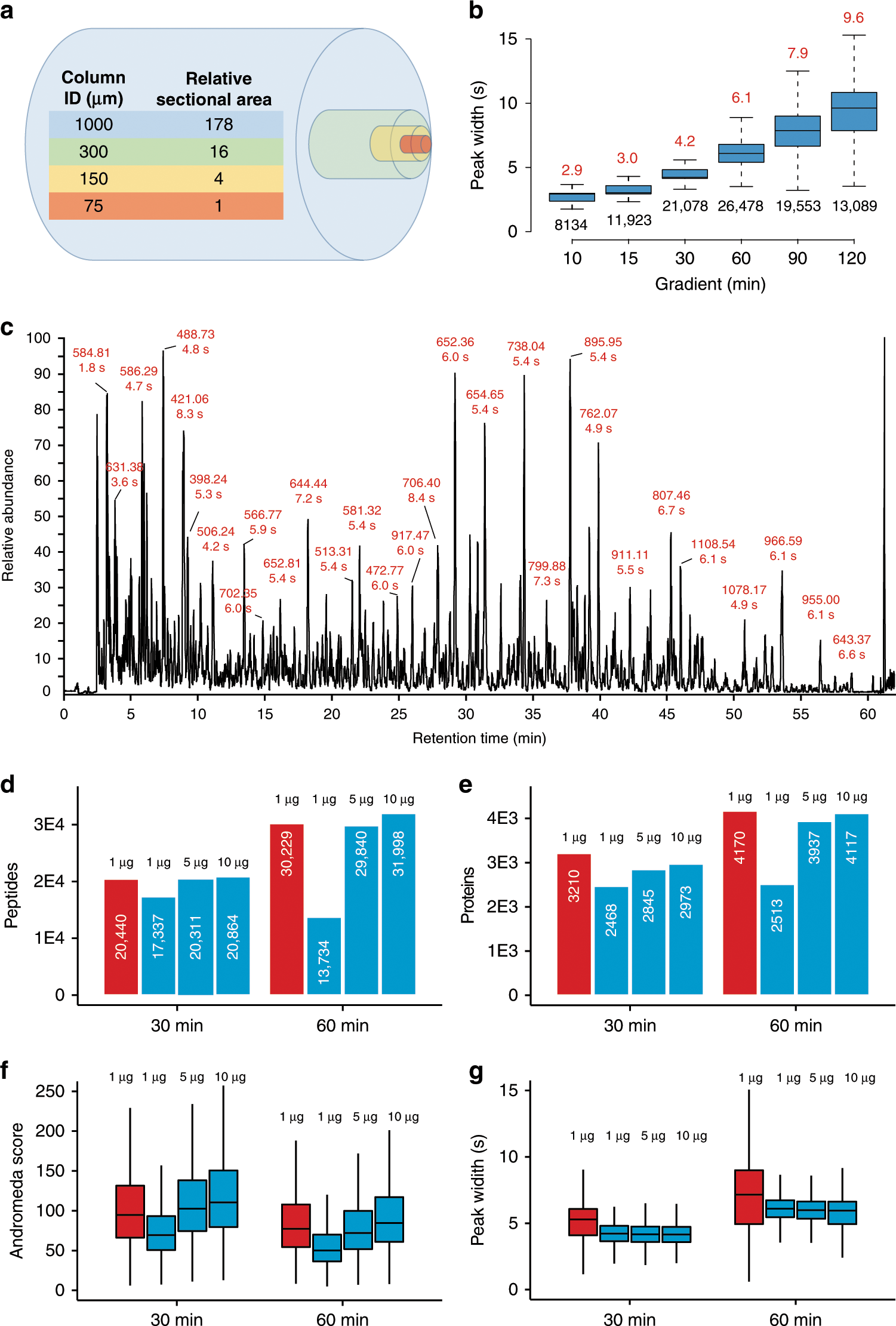
Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC–MS/MS | Nature Communications
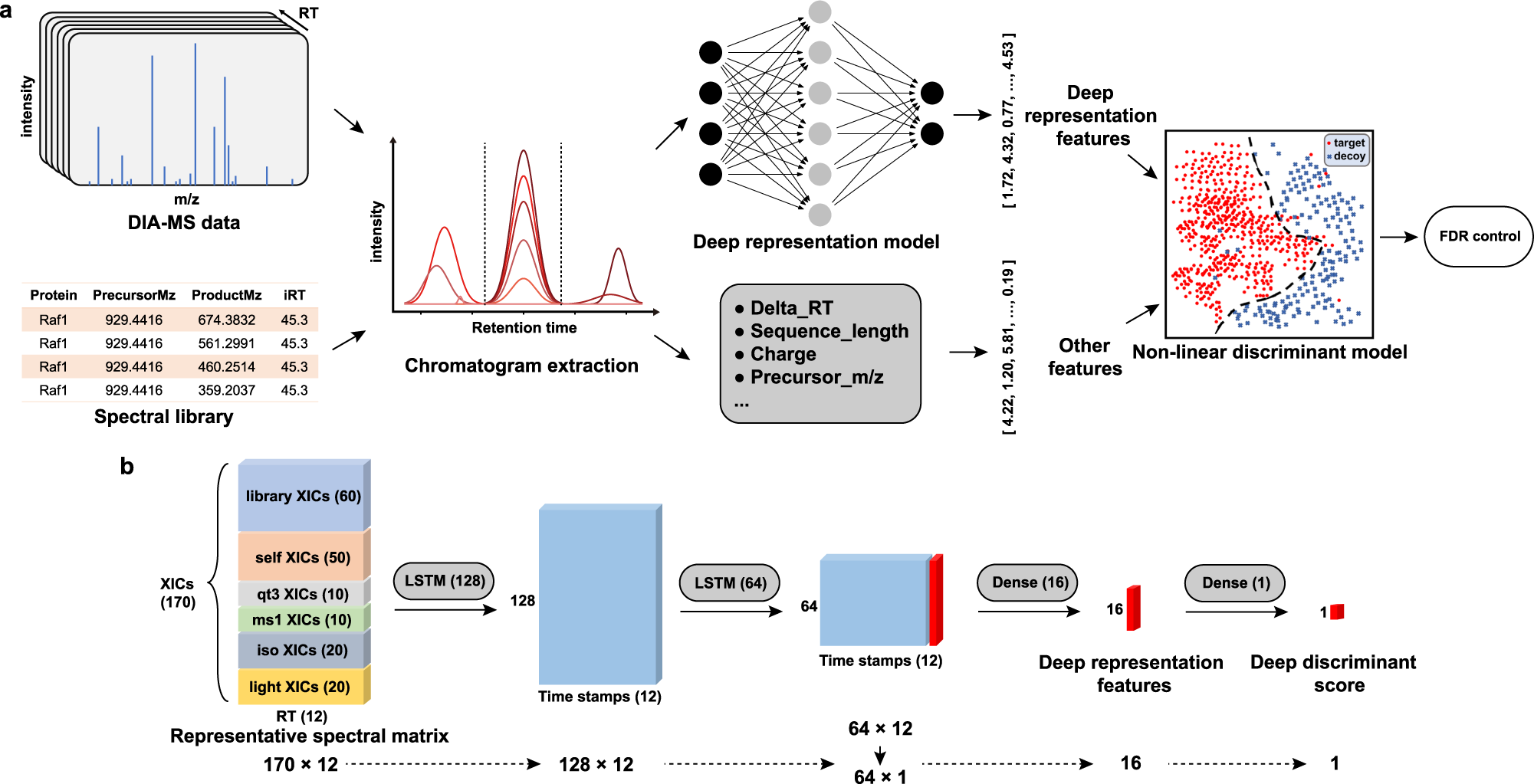
Deep representation features from DreamDIAXMBD improve the analysis of data-independent acquisition proteomics | Communications Biology
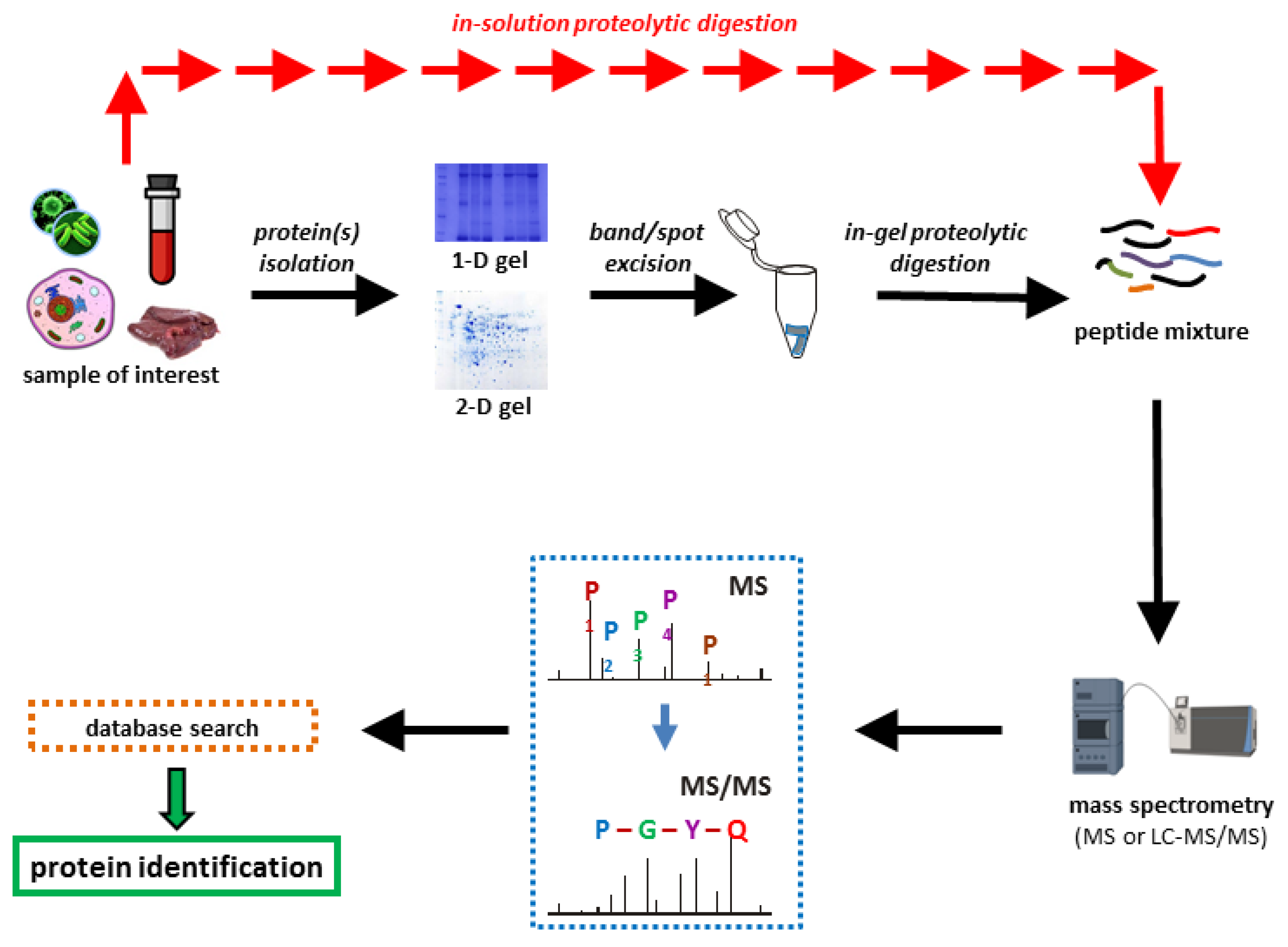
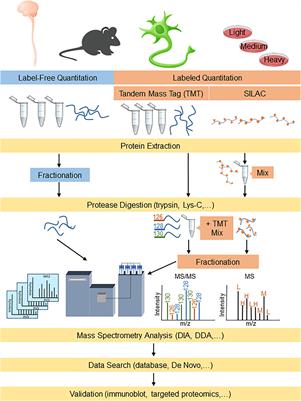
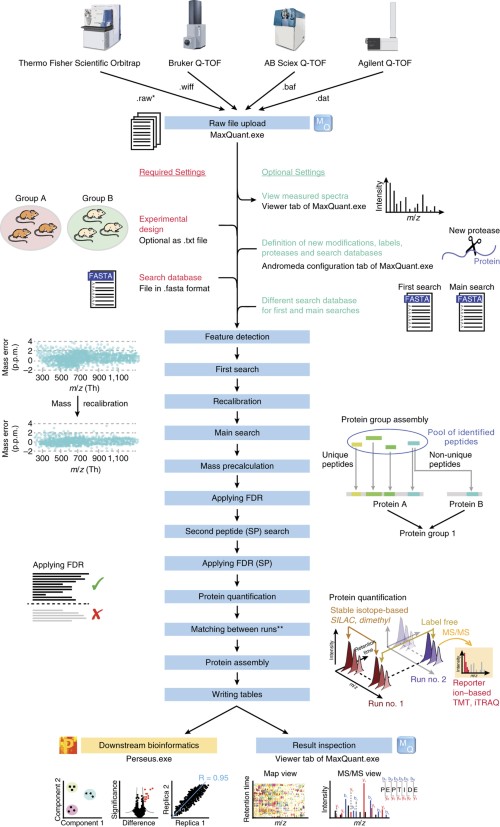
Proteomes | Free Full-Text | A Critical Review of Bottom-Up Proteomics: The Good, the Bad, and the Future of This Field
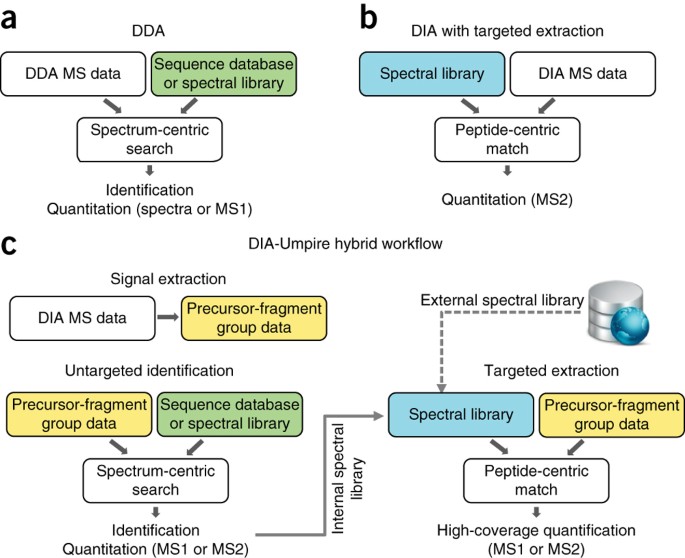
DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics | Nature Methods